Scanning Probe Lithography: DPN, PPL, µCS and FluidFM
Scanning probe lithography (SPL) methods like dip-pen nanolithography (DPN) [1], polymer pen lithography (PPL) [2], microcontactspotting (µCS) [3] and FluidFM [4] have all unique strengths in terms of resolution, obtainable throughput and patterning speed. Generally, SPL methods utilize the excellent control over positioning and movement of a tip (the probe) or sample stage as offered by atomic force microscopy (AFM) based technologies for highly localized deposition of “inks” (chemical compounds or carrier fluids with functional components). They work additive, maskless and exhibit broad compatibility with delicate chemical and biological inks, offer mild process parameters and are capable of multiplexing (i.e. deposition of different compounds within a desired micropattern) [5-7]. µCS and DPN in particular have shown great potential for low-volume liquid deposition in the femto- and attoliter regime for chemical surface functionalization [8]. The discussed SPL methods can functionalize surfaces over a wide range of resolutions and feature sizes. They can also be used for targeted functionalization of devices and pre-existing structures, e.g. for integration into microfluidics [9,10] or sensor functionalization [11-14], or do in-situ chemical synthesis [15].
| Name | Phone | |
|---|---|---|
| Dr. Dr. Dipl.-Phys. Michael Hirtz | +49 721 608-26373 | michael hirtz ∂does-not-exist.kit edu |
| Hussain | navid hussain ∂does-not-exist.kit edu |
Details (DPN)
Features
- Achievable resolution depends highly on ink/substrate combination. Highest resolution if offered by DPN (20 nm resolution for thiols on gold, or 100 nm for phospholipids), PPL and FluidFM offer high to medium resolution (50nm to 5µm) and µCS larger features (5µm to 50µm).
- Throughput on the order of cm2/min using massively parallel arrays
- Compatible with biological molecules (e. g. DNA, protein & phospholipids)
- Phospholipid based inks can write on a variety of surfaces – metals, insulators, hydrophobic, hydrophilic, etc.
- Capability of integrating multiple ink materials on a single substrate (Multiplexing)
- Compatible with pre-structured surfaces and pre-manufactured devices
- Additive fabrication process, digital and maskless
Limitations/constraints
- There must be a driving force for the ink to flow from the tip to the sample, stability of patterns depends on surface and ink properties and whether covalent binding occurs
- Parallel integration of different inks requires that the different inks have similar transport properties
- Each tip in a passive parallel array draws the same structure
- A high throughput quality control method must be used for massively parallel fabrication
- Area of single sub-pattern (before parallel repetition) is usually in the range of 60 × 60 to 100 × 100 µm2 depending on technology and machine
- Alignment marks must be used to align with pre-patterned substrates, prestructures must be observable by optical microscopy
- Standard sample size are aprox. 5 × 5 mm² to 2 × 2 cm², thicknesses about 0.5 to 2 mm. Samples of larger or smaller dimensions can be accommodated with special precautions.
Materials
A wide range of materials as inks and substrates can, be processed with the techniques [16,17], here some typical applications, please discuss your particular wish with the technology expert. In general:
- DPN, PPL: Molecular inks (e.g. thiols, azides, phospholipids), viscous inks (polymers, e.g. PEG)
- µCS, FluidFM: viscous inks (solvents carrying functional molecules or nanoparticles)
- Typical substrates: glass, silicon, PMMA, polystyrene, Metals (e.g. Au, Ti)
Typical structures and designs
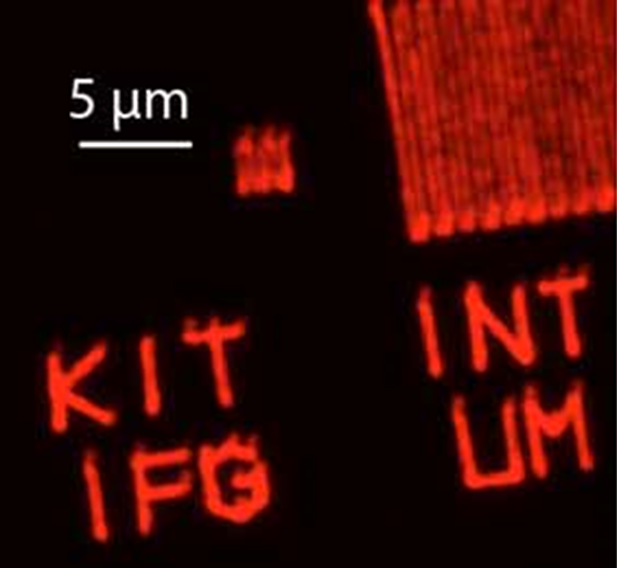
Fig. 1: Covalently linked fluorescently labeled azide ink written on an alkyne functionalized CVD coating [18]
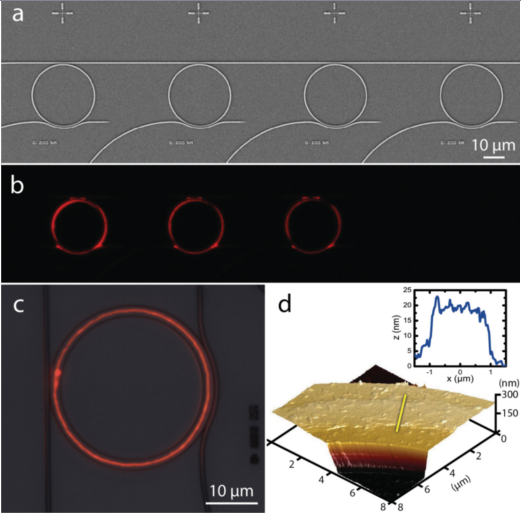

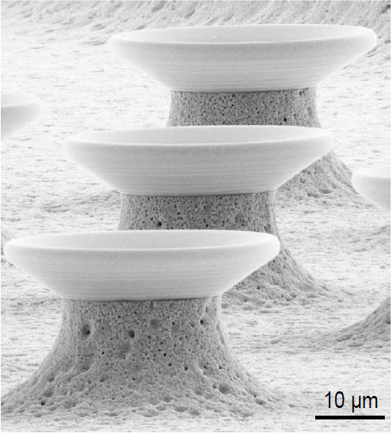
Fig. 2: Functionalization of pre-existing structures by DPN with lipids; photonic structures (top) [14], goblet structured sensors [11]
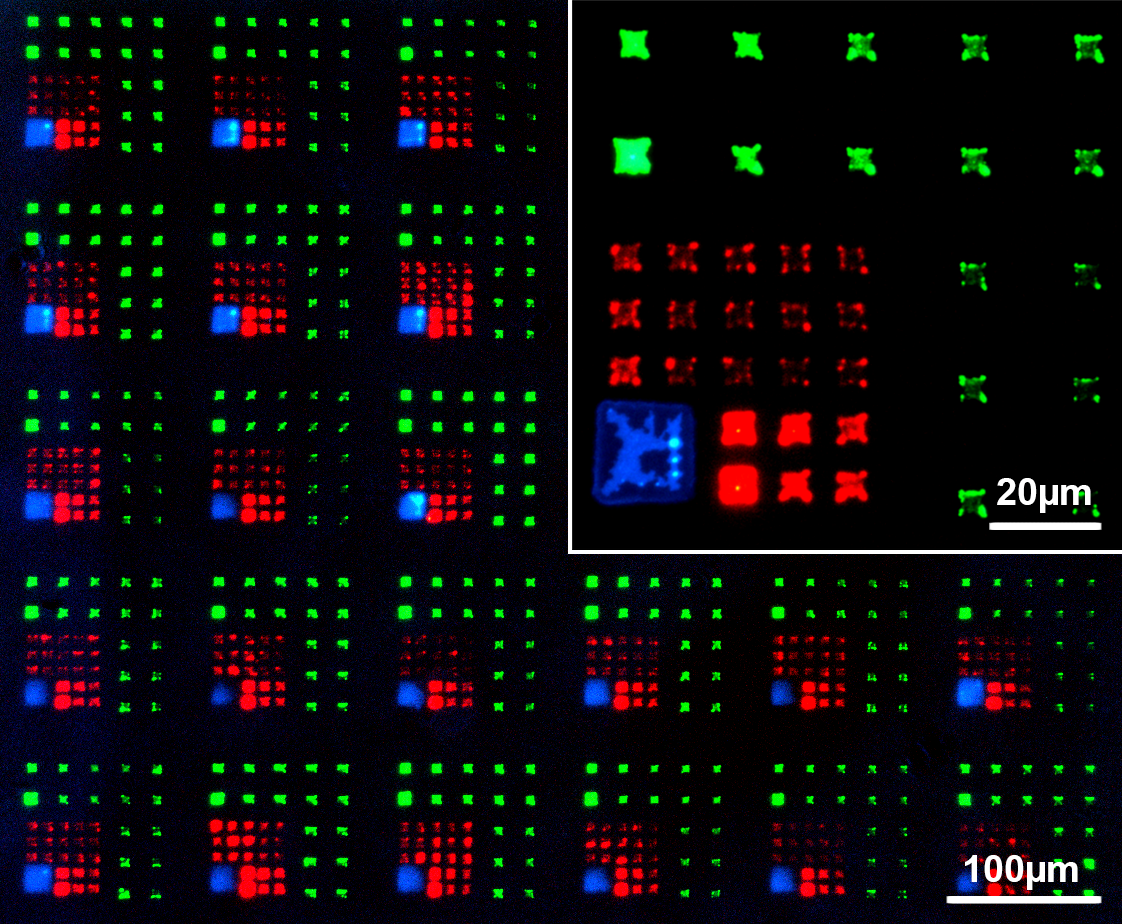
Fig. 3: Multi-color micro patterns generated by PPL with fluorescent lipids [19]
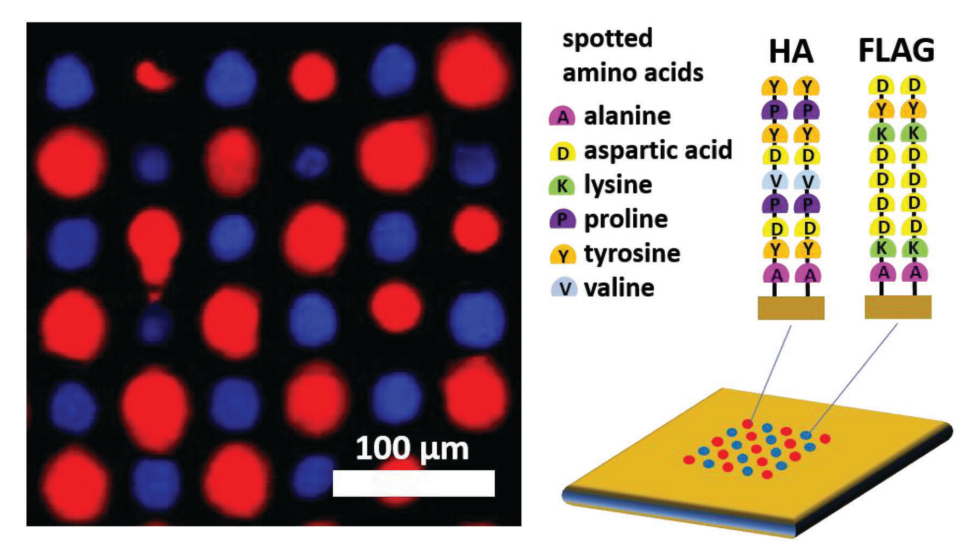
Fig. 4: A multiplexed array of in-situ synthesized peptide sequences [15]
References
[1] R. D. Piner, J. Zhu, F. Xu, S. Hong, C. A. Mirkin, Science 1999, 283, 661–663.
[2] F. Huo, Z. Zheng, G. Zheng, L. R. Giam, H. Zhang, C. A. Mirkin, Science 2008, 321, 1658–60.
[3] M. Hirtz, A. M. Greiner, T. Landmann, M. Bastmeyer, H. Fuchs, Adv. Mater. Interfaces 2014, 1, 1300129.
[4] A. Meister, M. Gabi, P. Behr, P. Studer, J. Vörös, P. Niedermann, J. Bitterli, J. Polesel-Maris, M. Liley, H. Heinzelmann, T. Zambelli, Nano Lett. 2009, 9, 2501–2507.
[5] G. Liu, M. Hirtz, H. Fuchs, Z. Zheng, Small 2019, 15, 1900564.
[6] A. Urtizberea, M. Hirtz, H. Fuchs, Nanofabrication 2015, 2, 43–53.
[7] H. Hu, H. Kim, S. Somnath, Micromachines 2017, 8, 90.
[8] M. Hirtz, S. Varey, H. Fuchs, A. Vijayaraghavan, ACS Appl. Mater. Interfaces 2016, 8, 33371–33376.
[9] F. Brinkmann, M. Hirtz, A. Haller, T. M. Gorges, M. J. Vellekoop, S. Riethdorf, V. Müller, K. Pantel, H. Fuchs, Sci. Rep. 2015, 5, 15342.
[10] R. Kumar, A. Bonicelli, S. Sekula-Neuner, A. C. B. Cato, M. Hirtz, H. Fuchs, Small 2016, 12, 5330–5338.
[11] U. Bog, T. Laue, T. Grossmann, T. Beck, T. Wienhold, B. Richter, M. Hirtz, H. Fuchs, H. Kalt, T. Mappes, Lab Chip 2013, 13, 2701–2707.
[12] U. Bog, F. Brinkmann, H. Kalt, C. Koos, T. Mappes, M. Hirtz, H. Fuchs, S. Köber, Small 2014, 10, 3863–3868.
[13] U. Bog, F. Brinkmann, S. F. Wondimu, T. Wienhold, S. Kraemmer, C. Koos, H. Kalt, M. Hirtz, H. Fuchs, S. Koeber, T. Mappes, Adv. Sci. 2015, 2, 1500066.
[14] P. Rath, M. Hirtz, G. Lewes-Malandrakis, D. Brink, C. Nebel, W. H. P. Pernice, Adv. Opt. Mater. 2015, 3, 328–335.
[15] J. Atwater, D. S. Mattes, B. Streit, C. von Bojničić-Kninski, F. F. Loeffler, F. Breitling, H. Fuchs, M. Hirtz, Adv. Mater. 2018, 30, 1801632.
[16] S. M. M. Dadfar, S. Sekula-Neuner, U. Bog, V. Trouillet, M. Hirtz, Small 2018, 14, 1800131.
[17] S. M. M. Dadfar, S. Sekula-Neuner, V. Trouillet, M. Hirtz, Adv. Mater. Interfaces 2018, 5, 1801343.
[18] H.-Y. Chen, M. Hirtz, X. Deng, T. Laue, H. Fuchs, J. Lahann, J. Am. Chem. Soc. 2010, 132, 18023–18025.
[19] F. Brinkmann, M. Hirtz, A. M. Greiner, M. Weschenfelder, B. Waterkotte, M. Bastmeyer, H. Fuchs, Small 2013, 9, 3266–3275.

